
Evolutionary Algorithms & Neuroevolution
Clear, modular & accessible evolution in Python
EvoLib is a lightweight, modular framework for evolutionary computation — built for research and teaching
pip install evolibfrom evolib import Population
pop = Population("examples/01_basic_usage/population.yaml")
pop.run()Why EvoLib?
Modular by Design
EvoLib combines classical evolutionary strategies (μ+λ, adaptive mutation, crossover) with structural neuroevolutioin a clear and modular frameworkrk.
Config-Driven Experiments
Define experiments via YAML with automatic validation - explicit, reproducible, and easy to adapt. Swap strategies or structures without touching core code.
Neuroevolution Built-in
Evolve weights, structure (connections, neurons, recurrence), and activations - combined with classical ES/GA methods.
Key features
Config-Driven Experiments
Define runs via YAML with automatic validation - no code changes needed.
Clear & Accessible
Clean API, transparent configs, examples that are easy to follow.
Classical Strategies
(μ, λ), (μ+λ), steady-state; tournament/roulette/rank/SUS selection.
Adaptive Operators
Constant, exponential decay, global & individual adaptation.
Neuroevolution
Evolve neural networks (EvoNet, structural growth).
Open & Extensible
MIT-licensed, flat API (from evolib import ...), easy to extend.
Quickstart
1) Setup
Install
pip install evolibCreate quickstart.yaml
# quickstart.yaml
parent_pool_size: 10
offspring_pool_size: 30
max_generations: 20
num_elites: 1
random_seed: 42
evolution:
strategy: mu_plus_lambda
modules:
main:
type: vector
dim: 8
bounds: [-1.0, 1.0]
initializer: random_vector
mutation:
strategy: constant
probability: 1.0
strength: 0.05
2) Create Python Script run_quickstart.py
# run_quickstart.py
import numpy as np
from evolib import Indiv, Population, sphere, plot_fitness
def my_fitness(indiv: Indiv) -> None:
x = indiv.para["main"].vector
indiv.fitness = sphere(x)
pop = Population("quickstart.yaml", fitness_function=my_fitness)
pop.run(verbosity=1)
# Basic plotting (default metrics: best, mean, and median fitness)
plot_fitness(pop, show=True)
3) Run
python run_quickstart.pyShowcases
Neuroevolution
Evolving network topology (add neurons/edges). Small, didactic, visual.
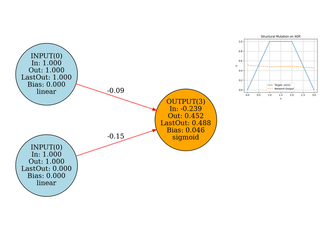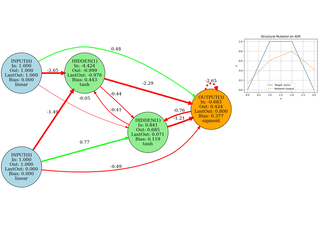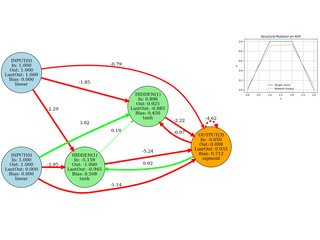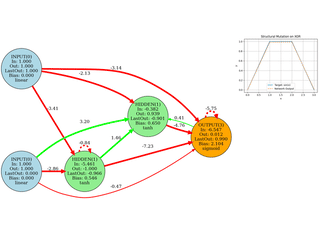
Image Approximation
Evolving an EvoNet to reproduce a target image. Useful for illustrating evolutionary concepts.
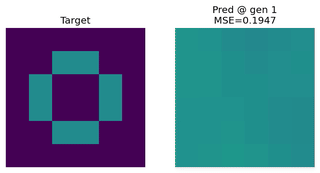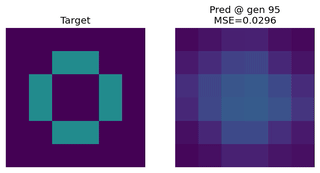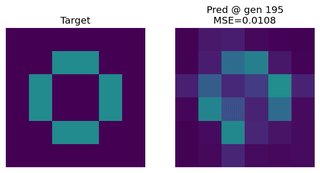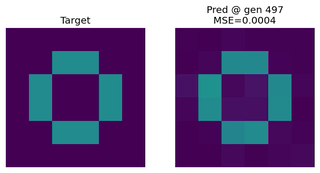
Function Approximation
Classic continuous benchmark with adaptive mutation strategies.

Extensions & Integrations
Parallel Evaluation with Ray
Seamless parallelization via Ray enables distributed fitness evaluation across CPU cores or clusters.
parallel:
backend: ray
num_cpus: 8
If no parallel block is given, EvoLib runs sequentially by default.
Gymnasium Integration
Native GymEnv wrapper for Gymnasium environments:
evolve neural controllers for tasks like CartPole, LunarLander, or BipedalWalker.
Explore available environments at
gymnasium.farama.org
.
from evolib import GymEnv
env = GymEnv("LunarLander-v3", max_steps=500)
fitness = env.evaluate(indiv, module="brain")Visualize your best agents easily with env.visualize()